
Eric Vitriol, PhD
Associate Professor
1120 15th St.
Room CA-3012
Augusta, GA 30912
office| 706-721-1176
lab| 706-721-1175
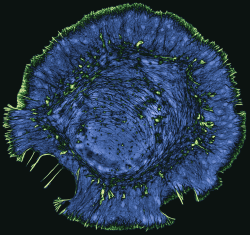
My lab studies the role of the actin cytoskeleton in cell motility, neural development, and in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS). We use live-cell and super-resolution imaging to understand how the dynamic regulation of actin contributes to the normal function of healthy cells, and how defects in actin can cause toxicity and cell death. We also benefit greatly from collaborations with computational groups, who help us build mathematical models and develop new methods of image analysis so that we can get the most out of our microscopy data. Our broad goal is to integrate imaging-based cell biology with translational science to understand how cell-based processes contribute to human disease.
We celebrate the strengths of people from diverse backgrounds and encourage those underrepresented in STEM to apply. If you are pursuing a research opportunity, please include a current CV and a brief statement about why the Vitriol lab is of interest in the initial email.
Postdoctoral researcher: We currently have one open postdoctoral position to investigate the cellular mechanisms of motor neuron degeneration in amyotrophic lateral sclerosis. This project will be highly microscopy-intensive, involving significant use of live-cell and super-resolution imaging, as well as quantitative image analysis. Experiments will be performed using immortalized cell lines, cultured mouse motor neurons, iPSC-derived human motor neurons, and patient samples from the local ALS clinic. Apply directly here (Job ID# 237445).
Graduate students: We welcome potential graduate students to apply for a rotation. Students are accepted through the Graduate Program in Neuroscience.
Undergraduate students: We accept undergraduate students who are interested in doing research for course credit through CURS. Must be willing to commit at least 8 hours per week in the lab. No previous experience needed- we will teach you everything that you need to know!
Deciphering the Mechanisms of Monomer-Driven Actin Dynamics
To divide, move, and communicate, cells rely on a dynamic actin cytoskeleton that
can rapidly assemble and change. This is achieved by the polymerization of actin monomers
into filaments, the construction of large filament networks, and the disassembly of
these networks back into monomers. Because it is so large, the monomer pool has traditionally
been considered to be homogeneous. However, recent work by us and others identified
distinct groups of monomers that drive and modify actin dynamics in ways that profoundly
influence cell behavior, revealing that the rules of monomer polymerization are much
more complex than previously thought. We are now developing approaches that merge
biochemical principles and microscopy to address these fundamental knowledge gaps
about monomer-driven actin dynamics. This includes controlling protein levels with
micromolar precision, using quantitative image analysis to extract rate constants
and concentrations, and employing computational models to reveal how cellular geometry
influences actin dynamics.
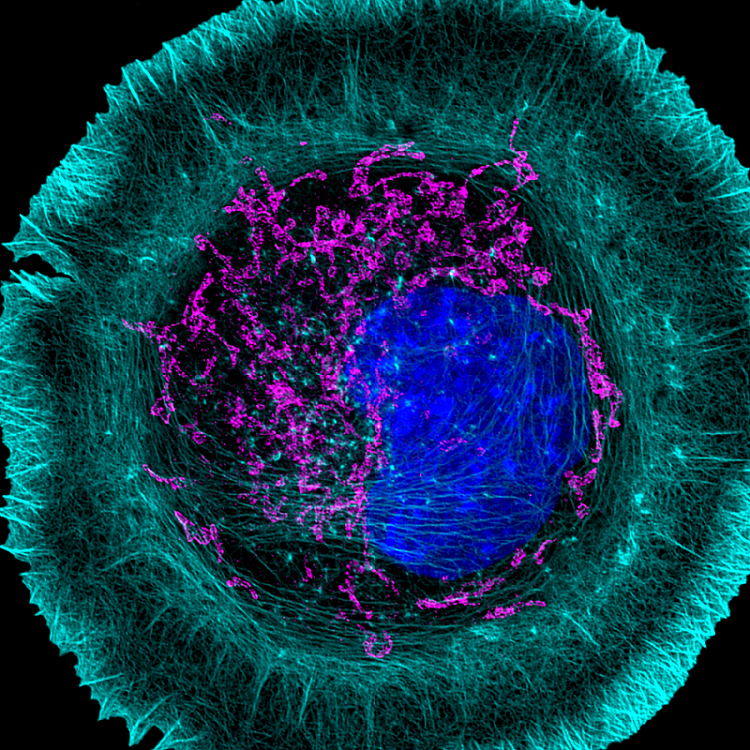
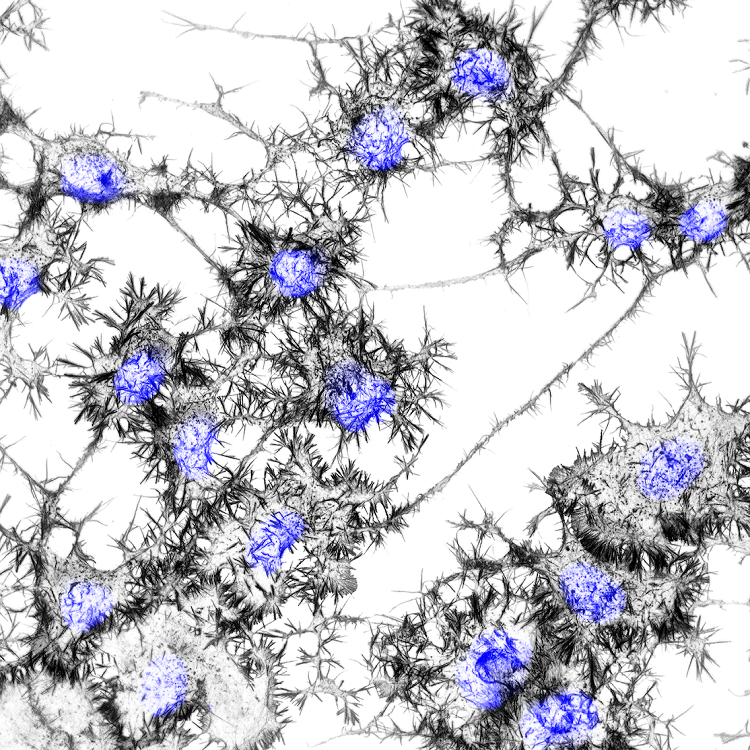
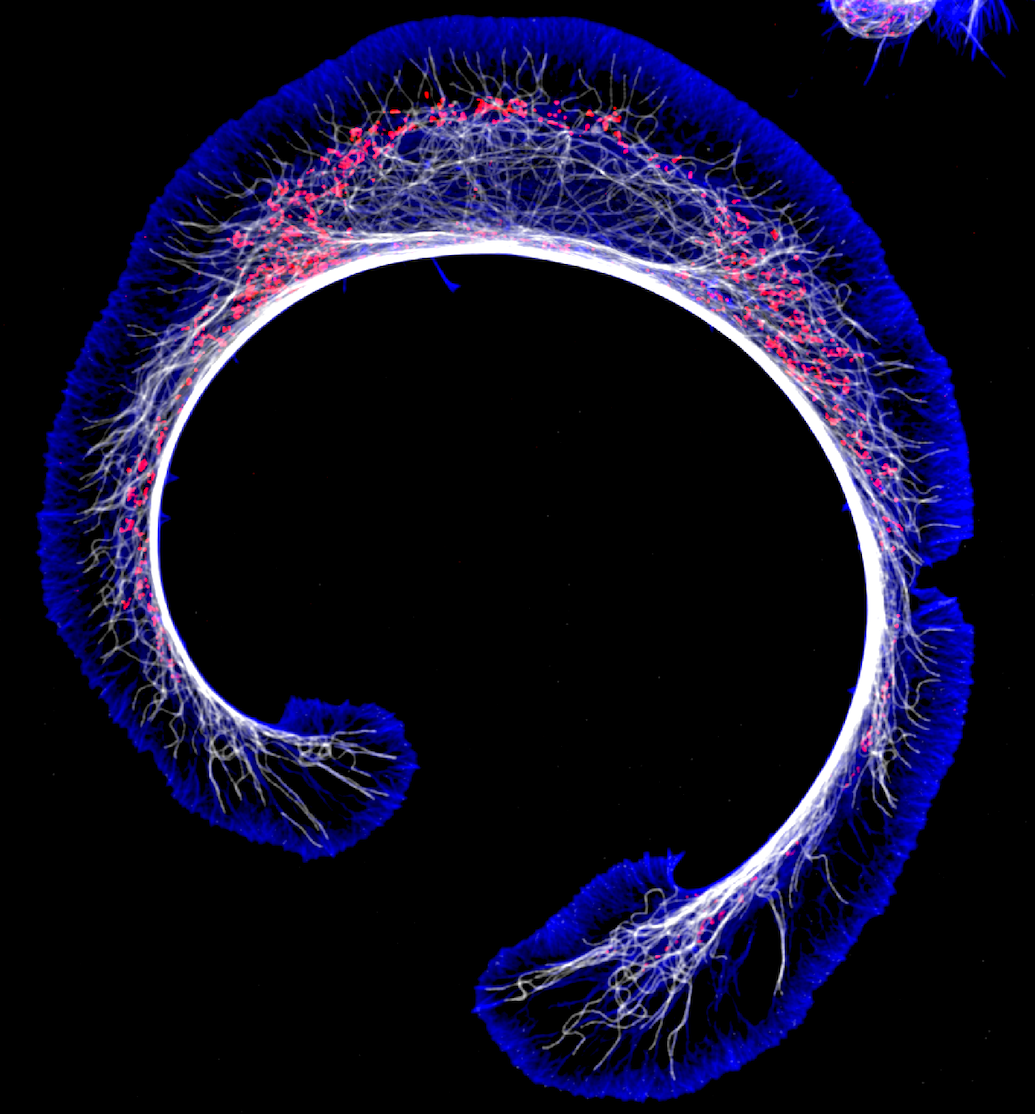
The Role of PFN1 in Motor Neuron Development and Disease
Profilin 1 (PFN1), the most prevalent monomer-binding protein in most mammalian cells,
prevents spontaneous actin polymerization and facilitates the assembly of specific
actin networks. We have recently shown that the precise control of PFN1 expression
levels defines which types of actin structures assemble from the monomer pool. PFN1
is important for neural development and regeneration and mutations in PFN1 cause a
hereditary form of the motor neuron disease amyotrophic lateral sclerosis (ALS). Using
live cell and super-resolution imaging of cultured neurons, we are now working to
better understand how PFN1 regulates the motor neuron cytoskeleton and controls their
physiology. Additionally, we are also using motor neurons derived from ALS mouse models
and induced pluripotent stem cells (iPSCs) of ALS patients to determine if defects
in the regulation of actin contribute to motor neuron disease.
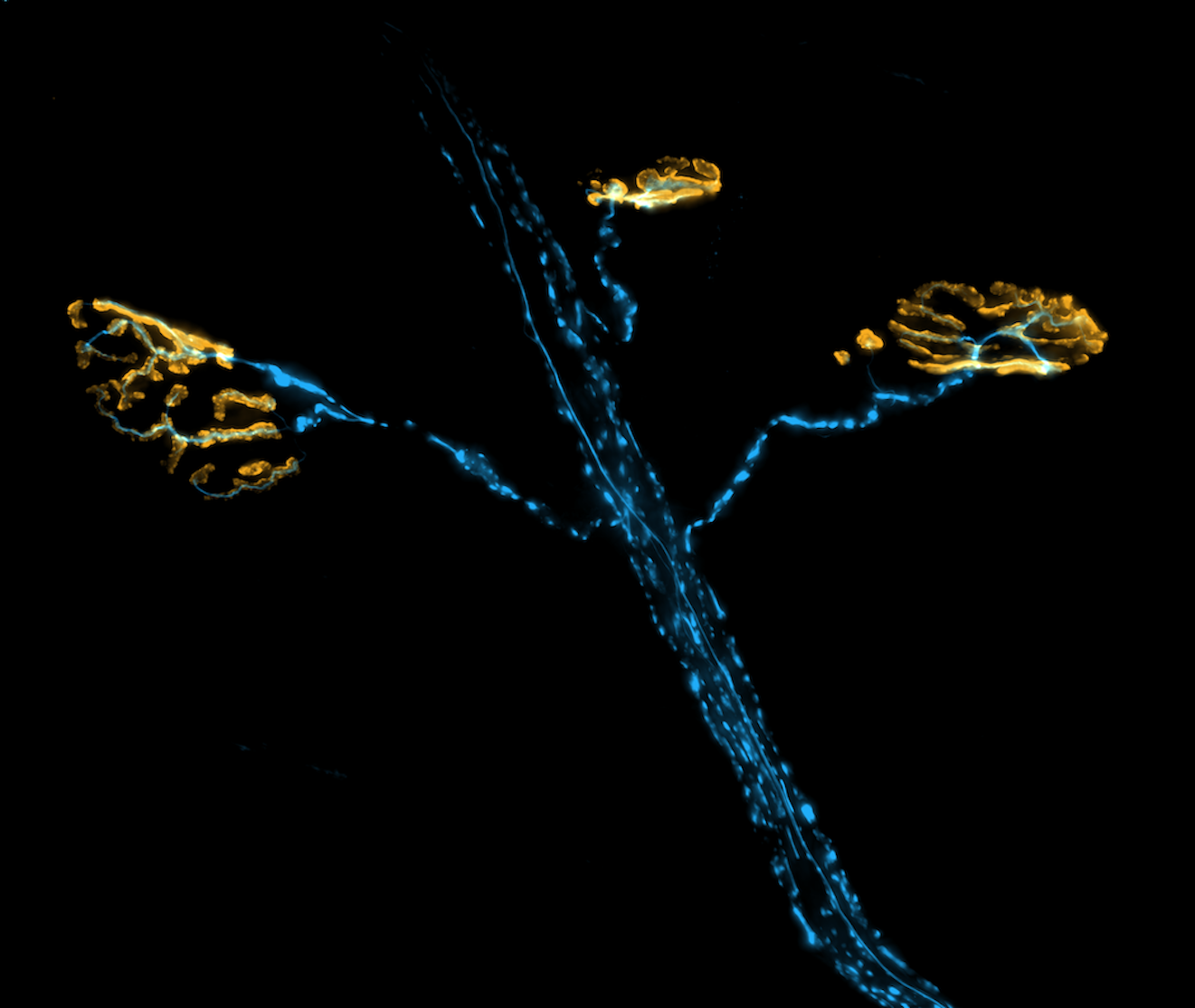
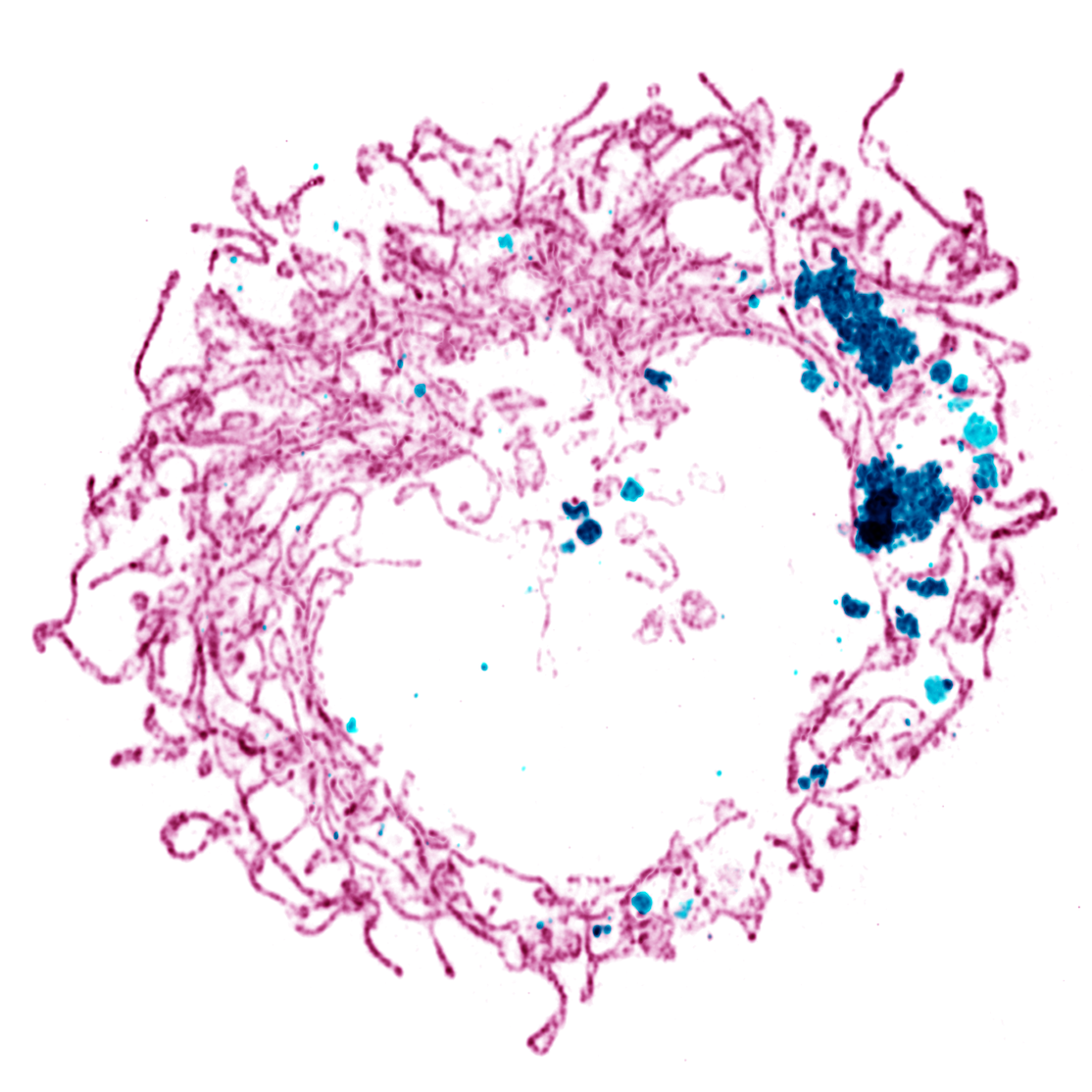
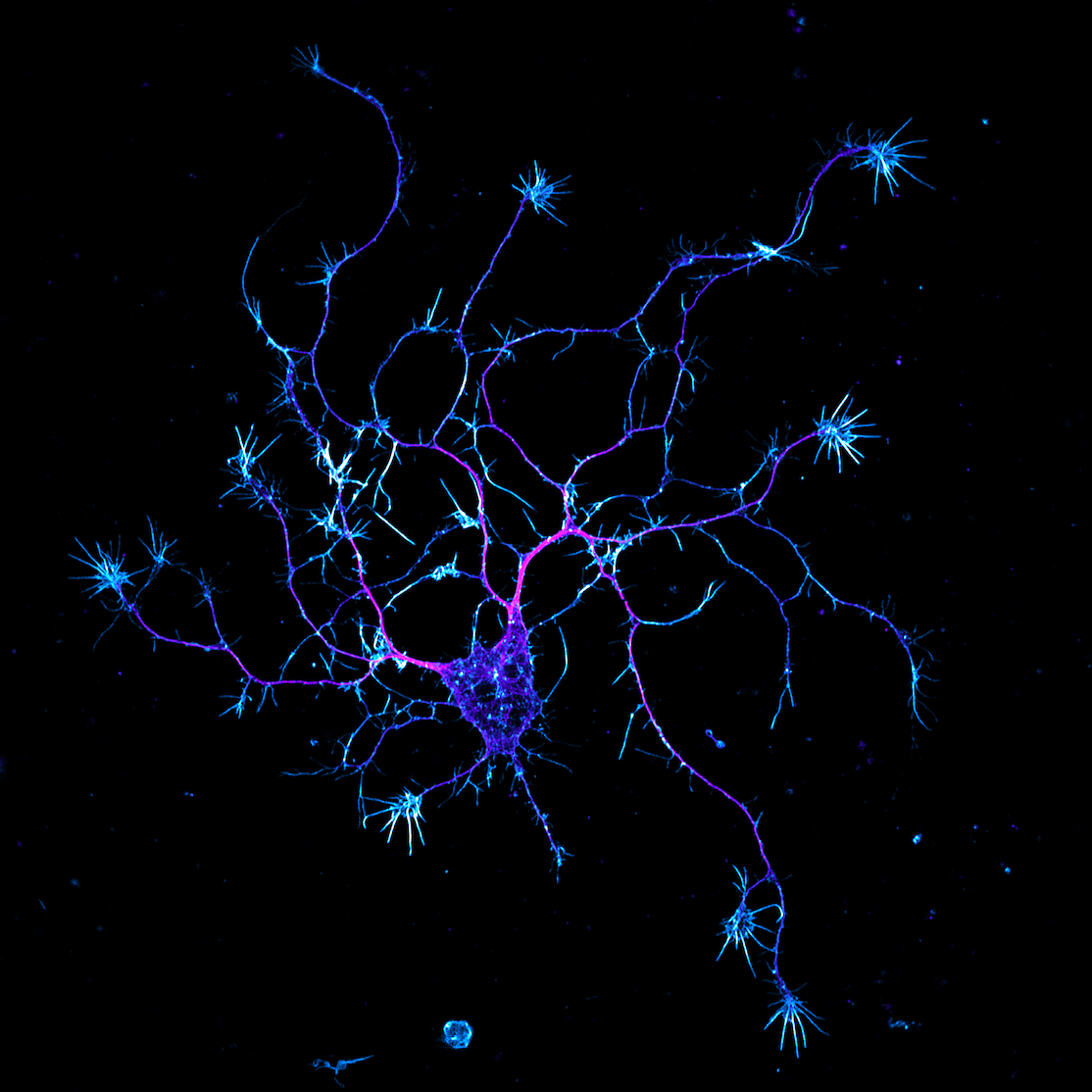
Edwards P, Skruber K, Milićević N, Heidings JB, Read TA, Bubenik P, Vitriol EA. (2021) TDAExplore: quantitative analysis of fluorescence microscopy images through topology-based machine learning. Patterns, 2(11): 100367. Link
Skruber K, Read TA, Vitriol EA. (2021) Delivering Defined Amounts of Purified Protein with High Precision into Living Cells. STAR Protocols, 2(1): 100272 Link
Skruber K, Warp PV, Shklyarov R, Thomas JD, Swanson MS, Henty-Ridilla JL, Read TA, Vitriol EA. (2020) Arp2/3 and Mena/VASP Require Profilin 1 for Actin Network Assembly at the Leading Edge. Current Biology, 30(14):2651-2664. Link
Osking Z, Ayers JI, Hildebrandt R, Skruber K, Eukovich AR, Brown H, Ryu D, Golde TE, Borchelt DR, Read TA, Vitriol EA. (2019) ALS-linked SOD1 Mutants Enhance Outgrowth and Branching in Adult Motor Neurons. iScience 11:294-304. Link
Skruber K, Read TA, Vitriol EA (2018) Reconsidering an Active Role for G-actin in Cytoskeletal Regulation. Journal of Cell Science, 131(1): jcs203760. Link
Kapustina M, Read TA, Vitriol EA (2016) Simultaneous Quantification of Actin Monomer and Filament Dynamics with Modelling Assisted Analysis of Photoactivation. Journal of Cell Science, 129(24):4633-4643. Link
Vitriol EA*, McMillen LM, Kapustina M, Gomez SM, Vavylonis D, Zheng JQ (2015) Two Functionally Distinct Sources of G-actin Supply the Leading Edge of Lamellipodia. Cell Reports, 11(3): 433-445. *Corresponding author Link