Yutao Liu, MD, PhD, FARVO

Professor
Director, CBA Graduate ProgramCellular Biology and Anatomy
Center for Biotechnology and Genomic Medicine
Basic Science Co-Director
James & Jean Culver Vision Discovery Institute
Office: Carl T. Sanders Research and Education Building, CB1123
Phone: 706-721-2015
Lab: 706-721-4843
yutliu@augusta.edu
EducationSelected Honors & AwardsResearch Goals & ApproachesMembersPublications & News/Media
Education
1990-1995 MD Beijing Medical University, Beijing, China
1999-2001 MS Truman State University, Kirksville, MO, USA
2001-2006 PhD University of Tennessee, Knoxville, TN, USA
2006-2010 Postdoctoral Associate, Duke University Medical Center, Durham NC, USA
Selected Honors & Awards
2010
2010-2013
The Thomas R. Lee Award for National Glaucoma Research, American Health Assistance Foundation, Clarksburg, MA
2013
Annual Educator Award, Division of Medical Genetics, Department of Medicine, Duke University School of Medicine
2017-2019
2020
2021
2022-now
2023
2017-now
2024-now
Research Goals & Approaches
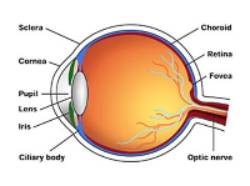
The long term goal of our laboratory is to dissect age-related genetic diseases (glaucoma and keratoconus) using systematic approaches, including human genetics/epigenetics, functional genomics, and molecular/cellular biology. Our lab has extensive research experience in the area of human genetics, bioinformatics, and functional genomics using several model systems, including human samples, in vitro cell/tissue culture and mouse models. Our research has been published in over 100 peer-reviewed articles, including Nature Genetics, PLoS Genetics, AJHG, PNAS, HMG, IOVS, EER, and Molecular Vision. Our research has been supported by a variety of different organizations, including the National Eye Institute at the National Institute of Health, several private foundations (The Glaucoma Foundation - TGF, the Glaucoma Research Foundation – GRF, the American Health Assistance Foundation – AHAF or BrightFocus, and RPB), and institutional supports.

Our current research is focused on understanding the molecular mechanisms and pathogenesis of two vision-related disorders: keratoconus (KC) and glaucoma. KC, a progressive thinning of the cornea, is the most common corneal ectasia, affecting one in every 500 to 2000 Americans. KC causes moderate to severe astigmatism, nearsightedness, swelling and cornea scarring. It is necessary for 10-20% KC patients to have cornea transplants. Using whole exome and whole genome sequencing, we have identified novel mutations in multiplex KC families. We are characterizing these mutations using primary human corneal cells and transgenic mouse models. We have research collaborations with investigators from California, Iowa, Illinois, Saudi Arabia, Israel, and Duke University in North Carolina. Glaucoma is the leading cause of irreversible blindness worldwide, affecting over 60 million people. It is characterized as a progressive loss of retinal ganglion cells and visual field.
Dr. Liu is a Co-Investigator of the NEIGHBORHOOD glaucoma genetics consortium.
Our lab has been involved in the identification of several glaucoma-associated genes, including but not limited to LOXL1, CDKN2B-AS1, SIX6, GALC, and chr8q22 locus. We are currently using primary human trabecular meshwork cells and other cells under cyclic mechanical stretch to determine their cellular functions in glaucoma. We are also interested in characterizing how the exosomes and its content – miRNA/proteins are involved in the pathogenesis of glaucoma and keratoconus. We have extensive experience in working with exosomes with > 30 peer-reviewed publications since 2013. Our team is highly motivated, cordial, collaborative, and trainee-friendly.
Approaches
We are using a number of advanced cutting-edge technologies, including whole exome/genome sequencing, Total RNA-Seq, small RNA-Seq, genome-wide DNA genotyping/gene expression, realtime PCR, droplet digital PCR, and nanoparticle tracking analysis (NTA) with ZetaView for exosomes and other nanoparticles. Bioinformatics is an important part of our research. We use a number of different bioinformatics tools in our daily research. We also use available mouse models to study both glaucoma and keratoconus by characterizing their ocular phenotypes.
Current & Former Members
Laboratory Members
Research Support Specialist: Hongfang Yu, B.S.Assistant Research Scientist: Jingwen Cai, M.S., PhD Graduate Student: Rachel Hadvina, B.S. Graduate Student: Ola Elsayed, B.S. Graduate Student: Mofazzal Hossain, B.S. Medical Student: Keerti Soundappan, B.S. Medical Student: Joshua Washington, B.S. Medical Student: Yejin (Jinny) Heo, B.S. Medical Student: Carol Beaty, B.S. Medical Student: Haley Chishom, B.S. Medical Student: Tarab Ajjan, B.S. Medical Student: Jason Sun, B.S.
Former Members
Graduate Student: Theresa Akoto, Ph.D. with Distinction (2023) Graduate Student: Hannah Youngblood, Ph.D. with Distinction (2022) Graduate Student: Jingwen Cai, M.S., Ph.D. (2019) Graduate Student: Mariam Khaled, M.S., Ph.D. (2019) Visiting Scholar: Yemin Zhang, Ph.D. Visiting Scholar: Yu Yan, Ph.D. STAR Student: Fikir Tefera, Lindsey Wilson College (2023) STAR Student: Hayden McCord, University of Georgia (2022) STAR Student: Jason Sun, University of Georgia (2021) STAR Student: Anush Aryal, Mississippi University for Women (2019) STAR Student: Rebecca Anderson, Emory University (2018) STAR Student: Sarah Yablonski, St. Lawrence University (2017) Postdoctoral Fellow: Inas Helwa, Ph.D.Postdoctoral Fellow: Mutsa Seremwe, Ph.D. Research Associate: Michelle Drewry, B.S. (2015-2018) Research Associate, Arthur Zimmerman, M.S. (2014-2015)
Medical Scholar (2015), Mohammed Waqar
Selected Recent Publications
| Publication |
|---|
Expression changes of human Schlemm's canal endothelial cells in response to cyclic mechanical stretchHeo, Y., Cai, J., Perkumas, K., Aboobakar, I. F., Stamer, W. D. & Liu, Y., Jan 2026, In: Experimental eye research. 262, 110747.Research output: Contribution to journal › Article › peer-review |
Proteomic Analysis Provides Insights Into PPIP5K2 Function and Its Impact on Corneal Energy MetabolismAkoto, T., Hao, C., Chen, Z., Lu, X., Yu, H., Gu, C., Shears, S. B., Zhi, W., Fan, X., Watsky, M. A. & Liu, Y., Dec 2025, In: Investigative Ophthalmology and Visual Science. 66, 15, 67.Research output: Contribution to journal › Article › peer-review |
Impact of psychosocial stress in early life on pace of aging in young adulthoodSu, S., Lewis, T. T., Belsky, D. W., Liu, Y., Zhang, K., Snieder, H. & Wang, X., Dec 2025, In: Clinical epigenetics. 17, 1, 186.Research output: Contribution to journal › Article › peer-review |
Plasma Extracellular Vesicle-Derived miR-296-5p is a Maturation-Dependent Rejuvenation Factor that Downregulates Inflammation and Improves Survival after SepsisCai, L., Kar, P., Liu, Y., Chu, X., Sharma, A., Lee, T. J., Arbab, A. & Raju, R. P., Apr 2025, In: Journal of Extracellular Vesicles. 14, 4, e70065.Research output: Contribution to journal › Article › peer-review |
Sexual dimorphism in the downregulation of extracellular matrix genes contributes to aortic stiffness in female miceKamau, A. N., Sakamuri, A., Okoye, D. O., Sengottaian, D., Cannon, J., Guerrero-Millan, J., Sullivan, J. C., Miller, K. S., Liu, Y. & Ogola, B. O., Mar 2025, In: American Journal of Physiology - Heart and Circulatory Physiology. 328, 3, p. H472-H483Research output: Contribution to journal › Article › peer-review |
Isolation and Characterization of Exosomes from Ocular SamplesCai, J. & Liu, Y., 2025, Methods in Molecular Biology. Humana Press Inc., p. 63-75 13 p. (Methods in Molecular Biology; vol. 2858).Research output: Chapter in Book/Report/Conference proceeding › Chapter |
Exfoliation syndrome genetics in the era of post-GWASElsayed, O. A., Cai, J. & Liu, Y., Jan 2025, In: Vision Research. 226, 108518.Research output: Contribution to journal › Article › peer-review |
Extracellular Vesicles and Glaucoma: Opportunities and ChallengesHossain, M. & Liu, Y., 2025, In: Current Eye Research. 50, 12, p. 1254-1263 10 p.Research output: Contribution to journal › Review article › peer-review |
Consensus Recommendations for Studies of Outflow Facility and Intraocular Pressure Regulation Using Ex Vivo Perfusion ApproachesAcott, T. S., Fautsch, M. P., Mao, W., Ethier, C. R., Huang, A. S., Kelley, M. J., Aga, M., Bhattacharya, S. K., Borras, T., Bovenkamp, D., Chowdhury, U. R., Clark, A. F., Dibas, M. I., Du, Y., Elliott, M. H., Faralli, J. A., Gong, H., Herberg, S., Johnstone, M. A. & Kaufman, P. L. & 49 others, Keller, K. E., Kelly, R. A., Krizaj, D., Kuehn, M. H., Li, H. L., Lieberman, R., Lin, S. C., Liu, Y., McDonnell, F. S., McDowell, C. M., McLellan, G. J., Mzyk, P., Nair, K. S., Overby, D. R., Peters, D. M., Raghunathan, V. K., Rao, P. V., Roddy, G. W., Sharif, N. A., Shim, M. S., Sun, Y., Thomson, B. R., Toris, C. B., Willoughby, C. E., Zhang, H. F., Freddo, T. F., Fuchshofer, R., Hill, K. R., Karimi, A., Kizhatil, K., Kopcyznski, C. C., Liton, P., Patel, G., Peng, M., Pattabiraman, P. P., Prasanna, G., Reina-Torres, E., Samples, E. G., Samples, J. R., Steel, C. L., Strohmaier, C. A., Subramanian, P., Sugali, C. K., van Batenburg-Sherwood, J., Wong, C., Youngblood, H., Zode, G. S., White, E. & Stamer, W. D., Dec 2024, In: Investigative Ophthalmology and Visual Science. 65, 14, 32.Research output: Contribution to journal › Article › peer-review |
A perspective from the National Eye Institute Extracellular Vesicle Workshop: Gaps, needs, and opportunities for studies of extracellular vesicles in vision researchLee, S. Y., Klingeborn, M., Bulte, J. W. M., Chiu, D. T., Chopp, M., Cutler, C. W., Das, S., Egwuagu, C. E., Fowler, C. D., Hamm-Alvarez, S. F., Lee, H., Liu, Y., Mead, B., Moore, T. L., Ravindran, S., Shetty, A. K., Skog, J., Witwer, K. W., Djalilian, A. R. & Weaver, A. M., Dec 2024, In: Journal of Extracellular Vesicles. 13, 12, e70023.Research output: Contribution to journal › Article › peer-review |
Influence of dexamethasone-induced matrices on the TM transcriptomeSoundappan, K., Cai, J., Yu, H., Dhamodaran, K., Baidouri, H., Vranka, J. A., Xu, H., Raghunathan, V. & Liu, Y., Nov 2024, In: Experimental eye research. 248, 110069.Research output: Contribution to journal › Article › peer-review |
Exploring New Links Among Keratoconus, Hormonal Factors, and Medications: Insights From a Case–Control Study Utilizing the All of Us DatabaseBeatty, C., Estes, A., Xu, H. & Liu, Y., Nov 2024, In: Translational Vision Science and Technology. 13, 11, 18.Research output: Contribution to journal › Article › peer-review |
Small Extracellular Vesicle-Associated MiRNAs in Polarized Retinal Pigmented EpitheliumHernandez, B. J., Strain, M., Suarez, M. F., Stamer, W. D., Ashley-Koch, A., Liu, Y., Klingeborn, M. & Rickman, C. B., Nov 2024, In: Investigative Ophthalmology and Visual Science. 65, 13, 57.Research output: Contribution to journal › Article › peer-review |
Pro-Inflammatory Characteristics of Extracellular Vesicles in the Vitreous of Type 2 Diabetic PatientsShan, S., Alanazi, A. H., Han, Y., Zhang, D., Liu, Y., Narayanan, S. P. & Somanath, P. R., Sep 2024, In: Biomedicines. 12, 9, 2053.Research output: Contribution to journal › Article › peer-review |
The impact of ischemic stroke on bone marrow microenvironment and extracellular vesicles: A study on inflammatory and molecular changesPatel, S., Khan, M. B., Kumar, S., Vyavahare, S., Mendhe, B., Lee, T. J., Cai, J., Isales, C. M., Liu, Y., Hess, D. C. & Fulzele, S., Sep 2024, In: Experimental Neurology. 379, 114867.Research output: Contribution to journal › Article › peer-review |
Evaluation of Novel Nasal Mucoadhesive Nanoformulations Containing Lipid-Soluble EGCG for Long COVID TreatmentFrank, N., Dickinson, D., Lovett, G., Liu, Y., Yu, H., Cai, J., Yao, B., Jiang, X. & Hsu, S., Jun 2024, In: Pharmaceutics. 16, 6, 791.Research output: Contribution to journal › Article › peer-review |
Identification of Keratoconus-Related Phenotypes in Three Ppip5k2 Mouse ModelsAkoto, T., Hadvina, R., Jones, S., Cai, J., Yu, H., McCord, H., Jin, C. X. J., Estes, A. J., Gan, L., Kuo, A., Smith, S. B. & Liu, Y., Jun 2024, In: Investigative Ophthalmology and Visual Science. 65, 6, 22.Research output: Contribution to journal › Article › peer-review |
Oral Microbially-Induced Small Extracellular Vesicles Cross the Blood-Brain BarrierElashiry, M., Carroll, A., Yuan, J., Liu, Y., Hamrick, M., Cutler, C. W., Wang, Q. & Elsayed, R., Apr 20 2024, In: International journal of molecular sciences. 25, 8Research output: Contribution to journal › Article › peer-review |
Feasibility Study of Developing a Saline-Based Antiviral Nanoformulation Containing Lipid-Soluble EGCG: A Potential Nasal Drug to Treat Long COVIDFrank, N., Dickinson, D., Garcia, W., Liu, Y., Yu, H., Cai, J., Patel, S., Yao, B., Jiang, X. & Hsu, S., Feb 2024, In: Viruses. 16, 2, 196.Research output: Contribution to journal › Article › peer-review |
Estrogen dysregulation, intraocular pressure, and glaucoma riskYoungblood, H., Schoenlein, P. V., Pasquale, L. R., Stamer, W. D. & Liu, Y., Dec 2023, In: Experimental eye research. 237, 109725.Research output: Contribution to journal › Review article › peer-review |
Animal Models for the Study of KeratoconusHadvina, R., Estes, A. & Liu, Y., Dec 2023, In: Cells. 12, 23, 2681.Research output: Contribution to journal › Review article › peer-review |
Mechanosensitive ion channel gene survey suggests potential roles in primary open angle glaucomaNEIGHBORHOOD Consortium, Dec 2023, In: Scientific reports. 13, 1, 15871.Research output: Contribution to journal › Article › peer-review |
Exosomes and their miRNA/protein profile in keratoconus-derived corneal stromal cellsHadvina, R., Lotfy Khaled, M., Akoto, T., Zhi, W., Karamichos, D. & Liu, Y., Nov 2023, In: Experimental eye research. 236, 109642.Research output: Contribution to journal › Article › peer-review |
Polarized desmosome and hemidesmosome shedding via small extracellular vesicles is an early indicator of outer blood-retina barrier dysfunctionHernandez, B. J., Skiba, N. P., Plössl, K., Strain, M., Liu, Y., Grigsby, D., Kelly, U., Cady, M. A., Manocha, V., Maminishkis, A., Watkins, T. J., Miller, S. S., Ashley-Koch, A., Stamer, W. D., Weber, B. H. F., Bowes Rickman, C. & Klingeborn, M., Oct 2023, In: Journal of Extracellular Biology. 2, 10, e116.Research output: Contribution to journal › Article › peer-review |
Glaucomatous aqueous humor vesicles are smaller and differ in composition compared to controlsMueller, A., Anter, A., Edwards, G., Junk, A. K., Liu, Y., Ziebarth, N. & Bhattacharya, S. K., Sep 2023, In: Experimental eye research. 234, 109562.Research output: Contribution to journal › Article › peer-review |
Genetic Correlations Among Corneal Biophysical Parameters and Anthropometric TraitsCousins, H. C., Cousins, C. C., Valluru, G., Altman, R. B., Liu, Y., Pasquale, L. R. & Ahmad, S., Aug 1 2023, In: Translational Vision Science and Technology. 12, 8, 8.Research output: Contribution to journal › Article › peer-review |
Engineered Human Dendritic Cell Exosomes as Effective Delivery System for Immune ModulationElsayed, R., Elashiry, M., Tran, C., Yang, T., Carroll, A., Liu, Y., Hamrick, M. & Cutler, C. W., Jul 2023, In: International journal of molecular sciences. 24, 14, 11306.Research output: Contribution to journal › Article › peer-review |
Mitochondrial TXNRD2 and ME3 Genetic Risk Scores Are Associated with Specific Primary Open-Angle Glaucoma PhenotypesNEIGHBORHOOD Consortium, Jul 2023, In: Ophthalmology. 130, 7, p. 756-763 8 p.Research output: Contribution to journal › Article › peer-review |
Homocysteine and GlaucomaWashington, J., Ritch, R. & Liu, Y., Jul 2023, In: International journal of molecular sciences. 24, 13, 10790.Research output: Contribution to journal › Review article › peer-review |
Src family kinases engage differential pathways for encapsulation into extracellular vesiclesYe, C., Gosser, C., Runyon, E. D., Zha, J., Cai, J., Beharry, Z., Bowes Rickman, C., Klingeborn, M., Liu, Y., Xie, J. & Cai, H., Jun 2023, In: Journal of Extracellular Biology. 2, 6, e96.Research output: Contribution to journal › Article › peer-review |
Unravelling the Impact of Cyclic Mechanical Stretch in Keratoconus—A Transcriptomic Profiling StudyAkoto, T., Cai, J., Nicholas, S., McCord, H., Estes, A. J., Xu, H., Karamichos, D. & Liu, Y., Apr 2023, In: International journal of molecular sciences. 24, 8, 7437.Research output: Contribution to journal › Article › peer-review |
Transcriptome Analysis of Retinal and Choroidal Pathologies in Aged BALB/c Mice Following Systemic Neonatal Murine Cytomegalovirus InfectionZhang, X., Xu, J., Marshall, B., Dong, Z., Liu, Y., Espinosa-Heidmann, D. G. & Zhang, M., Mar 2023, In: International journal of molecular sciences. 24, 5, 4322.Research output: Contribution to journal › Article › peer-review |
Electrical Stimulation Increases the Secretion of Cardioprotective Extracellular Vesicles from Cardiac Mesenchymal Stem CellsZhang, H., Shen, Y., Kim, I. M., Liu, Y., Cai, J., Berman, A. E., Nilsson, K. R., Weintraub, N. L. & Tang, Y., Mar 2023, In: Cells. 12, 6, 875.Research output: Contribution to journal › Article › peer-review |
Microbially-Induced Exosomes from Dendritic Cells Promote Paracrine Immune Senescence: Novel Mechanism of Bone Degenerative Disease in MiceElsayed, R., Elashiry, M., Liu, Y., Morandini, A. C., El-Awady, A., Elashiry, M. M., Hamrick, M. & Cutler, C. W., Feb 1 2023, In: Aging and Disease. 14, 1, p. 136-151 16 p.Research output: Contribution to journal › Article › peer-review |
Defining the ligand-dependent proximatome of the sigma 1 receptorZhao, J., Veeranan-Karmegam, R., Baker, F. C., Mysona, B. A., Bagchi, P., Liu, Y., Smith, S. B., Gonsalvez, G. B. & Bollinger, K. E., 2023, In: Frontiers in Cell and Developmental Biology. 11, 1045759.Research output: Contribution to journal › Article › peer-review |
Nebulization of extracellular vesicles: A promising small RNA delivery approach for lung diseasesHan, Y., Zhu, Y., Youngblood, H. A., Almuntashiri, S., Jones, T. W., Wang, X., Liu, Y., Somanath, P. R. & Zhang, D., Dec 2022, In: Journal of Controlled Release. 352, p. 556-569 14 p.Research output: Contribution to journal › Article › peer-review |
Revealing the presence of tear extracellular vesicles in KeratoconusHefley, B. S., Deighan, C., Vasini, B., Khan, A., Hjortdal, J., Riaz, K. M., Liu, Y. & Karamichos, D., Nov 2022, In: Experimental eye research. 224, 109242.Research output: Contribution to journal › Article › peer-review |
Identification of circulating microvesicle-encapsulated miR-223 as a potential novel biomarker for ARDSAlmuntashiri, S., Han, Y., Youngblood, H. A., Chase, A., Zhu, Y., Wang, X., Linder, D. F., Siddiqui, B., Sikora, A., Liu, Y. & Zhang, D., Nov 2022, In: Physiological reports. 10, 21, e15494.Research output: Contribution to journal › Article › peer-review |
Blunted rest-activity circadian rhythm is associated with increased rate of biological aging: an analysis of NHANES 2011-2014Xu, Y., Wang, X., Belsky, D. W., McCall, W. V., Liu, Y. & Su, S., Sep 18 2022, In: The journals of gerontology. Series A, Biological sciences and medical sciences.Research output: Contribution to journal › Article › peer-review |
The Underlying Relationship between Keratoconus and Down SyndromeAkoto, T., Li, J. J., Estes, A. J., Karamichos, D. & Liu, Y., Sep 2022, In: International journal of molecular sciences. 23, 18, 10796.Research output: Contribution to journal › Review article › peer-review |
For a detailed list of recent publications, please refer to NCBI My Bibliography
*** Please note that some of the listed publications were included as the NEI P30-supported studies but not as a co-author since Dr. Liu is the PI of the P30 grant. These should be treated as P30-supported publications ONLY.
News/Media Coverage
- Our research on exosome research with Particle Matrix’s ZetaView
- Three glaucoma-related genes discovered from Science Daily
- Liu’s interview with MD Magazine at the 2016 annual ARVO meeting at Seattle, WA
- Our research discovery of miR-182 association with POAG and funding from BrightFocus
by multiple media
JAGWIRE, Science Daily, MedicalXpress, BioQuick News - Dr. Liu’s participation of 2015 and 2016 Think Tank for The Glaucoma Foundation in New York City
- Our Aqueous humor miRNA screening published in Human Molecular Genetics 2018 by multiple media
Science Daily, Deccan Chronicle, EurekAlert, News Medical - Our research in the role of a long noncoding RNA in the LOXL1 locus in exfoliation
glaucoma by multiple media
Science Daily, Technology Networks